Enzyme kinetics
The study of enzyme kinetics is the ground of enzyme production for industry. Actually, a lot of Enzymes are applied and produced with mass production. Therefore, It is very important to understand enzyme reaction kinetics and the kinetics are based on the “Michaelis-Menten equation and Lineweaver-Burk graph“.
- Michaelis-Menten equation: V0 = Vmax * [s] / (km + [s])
- # Lineweaver-Burk plot: 1/V0 = km/(Vmax * [s]) + 1/Vmax
In this experiment, ß-amylase was used for converting starch into maltose. DNS reaction was performed to analyze maltose concentration by converting maltose to 3-amino-5-mitrusalicylic acid. This 3-amino-5-mitrusalicylic acid tends to absorb at 575 nm wavelength.
효소 동역학 연구는 산업용 효소 생산의 기초가 됩니다. 실제로 많은 Enzyme이 실생활에 적용되어 대량 생산되고 있습니다. 따라서 효소 반응 역학을 이해하는 것이 매우 중요하며, 이 역학은 “Michaelis-Menten 방정식과 Lineweaver-Burk 그래프”를 기반으로 합니다.
- Michealis-Menten equation: V0 = Vmax * [s] / (km + [s])
- # Lineweaver-Burk plot: 1/V0 = km/(Vmax * [s]) + 1/Vmax
이 실험에서, β-아밀레이스는 녹말을 말토스로 변환하는 데 사용되었습니다. DNS 반응은 말토스를 3-아미노-5-미트루살실산으로 변환하여 말토스 농도를 분석하기 위해 수행되었습니다. 이 3-아미노-5-미트루살실산은 575nm 파장에서 흡수되는 경향이 있습니다.
Materials
DNS reagent, Maltose solution, starch solution, beta-amylase, 0.1M NaOH solution, 0.1M Phosphate buffer
Pipette & tips, Timers, Spectrophotometer, Test tube
Methods
- Add the following reagents to each test tube with 1.25 mL of maltose solutions (0.0, 0.2, 0.4, 0.6, 0.8, & 1.0 mg/mL) and 1 mL of Phosphate buffer
- Take 2.25 mL of solution and mix with the same volume of DNS reagent
- Warm up the solution at 100℃ in a double boiler for 15 minutes and then cool at room temperature
- Measure the absorbance of each solution at 575 nm wavelength (If OD >1, dilute the solution up to 1/2~1/10)
- Add 1 mL Phosphate buffer and 1.25 mL starch solutions (5, 10, 15, & 20mg/mL) to the test tube and put in a 37℃ water bath for 10 minutes.
- Add 0.25 mL of ß-amylase to the test tube and stand for 2 minutes
- To inactivate the enzyme, add 5 mL of 0.1 N NaOH, and store at 4℃ for 5 minutes
- Get 3 mL of solution and mix it with the same volume of DNS reagent
- Warm up the solution at 100℃ in a double boiler for 15 minutes and then cool at room temperature
- Measure the absorbance of each solution at 575 nm wavelength (If OD >1, dilute the solution up to 1/2~1/10).
4. Result
A standard graph result
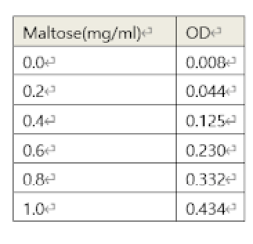
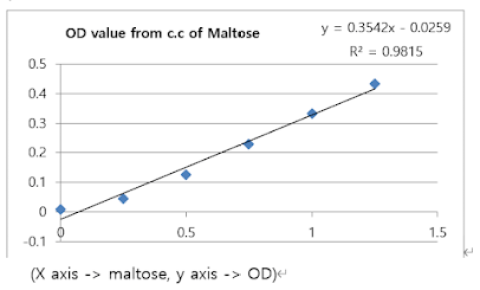
Certain concentrations of beta-amylase reacted with several concentrations of starch (5, 10, 15, 20) (mg/ml) for 2 minutes. After the DNS reaction of the samples, each OD value was measured at 0.022, 0.050, 0.092, and 0.138. With the standard’s graph equation (y=0.3542x – 0.0259; y->OD, x-> maltose), Maltose concentrations were calculated. The result is below in the table.

The starch was reacted for 2 minutes. So, V0 value is ‘maltose concentration/2’ (mg/ml*t). And 1/[s], and 1/V0 were calculated.

Based on this data, the plot (X-> 1/[s] / Y -> 1/V0) was drawn.
# Lineweaver-Burk plot: 1/V0 = km/(Vmax * [s]) + 1/Vmax
Y = 67.903x + 1.541 = 1/V0= 67.903 * 1/[s] + 1.541
Therefore, km/ Vmax = 67.903, and 1/Vmax is 1.541. V max is 0.649 mg/ml*min, and km is 44.075 mg/ml.
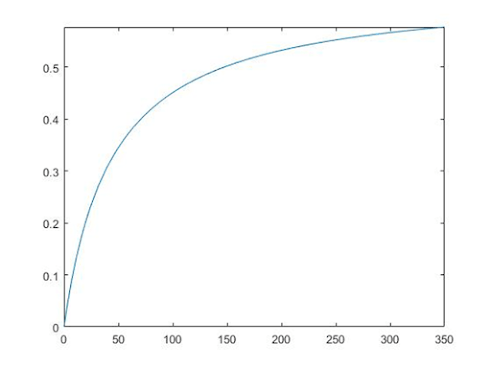
As a result, the Michaelis-Menten equation of the ß-amylase enzyme in this experiment is
V= 0.6491*[s]/(44.075 + [s]).
This graph is below. This graph is used in “Matlab” and [s] rage is 0~350 mg/ml.
5. Discussion
The experiment was good because all of the graph R2 values are not low, >0.98. Lastly, michaelis-menten’s graph also could be obtained.
(학사때 써논 건데 혹시나 필요하신 분들을 위한 정보입니다. 학사때 쓴거라 좀 허접함 주의)
'기타 등등' 카테고리의 다른 글
| 선넘게 비싼 굿노트6 사야할까? (반 값이면 바로 샀을 듯) (1) | 2023.11.20 |
|---|---|
| [생물공학실험] The basics of Animal cell culture and calculation doubling time (1) | 2023.10.01 |
| [생물공학실험] Gram-Staining (Gram-negative and -positive) with using Chemicals (0) | 2023.10.01 |
| [생물공학실험]Inoculation and Culture for Microbes (Streaking and Spreading) (0) | 2023.10.01 |
| [생물공학실험] E. Coli Culture, LB Media Preparation And Sterilization (0) | 2023.10.01 |




